Baird Lab
Manipulating proteases, so you don't have to. Welcome to the homepage of the Baird Laboratory at San Francisco State University in the Department of Chemistry and Biochemistry. Thanks for visiting our website. If you would like to know more about what we do, please send me an email. I’m happy to share!
Research
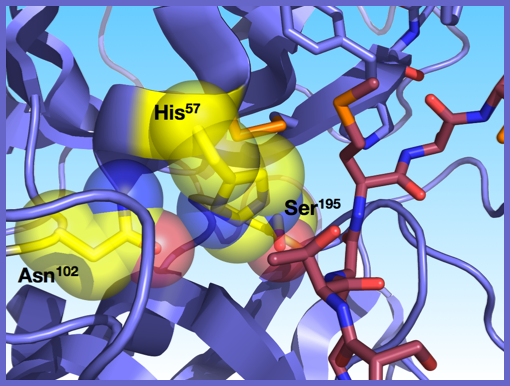
We are a biochemistry lab in the Chemistry and Biochemistry Department at San Francisco State University. Our research interests are in protein structure-function relationships, particularly substrate specificity, catalysis and inhibition in serine proteases. I have little doubt that you've heard of the serine protease trypsin. It's in every biochemistry text book, it's used in mass spectroscopy studies and it's been studied for over 50 years. Well, in the Baird Lab, we use trypsin as a model system because its scaffold is representative of a number of other serine proteases making our results potentially applicable to more than 100 other enzymes. Because there is so much already known about it, it an excellent system for protein engineering experiments in this family of enzymes and it is relatively easy to mutagenize, express, purify and characterize. A couple of the questions we are asking are…
One of the basic science questions we asked was, why do we not observe many threonine proteases in nature? Using molecular modeling, we hypothesized that structural limitations imposed by the presence of a disulfide bridge near the active site precluded the use of the chemically similar, but larger threonine at position 195. Consequently, we hypothesized that we could remove this disulfide bridge which should allow for threonine to be used similarly to serine in the catalytic mechanism. We were successful in creating a functional threonine protease within the trypsin scaffold. In collaboration with the Guliaev Lab at SF State, we carried out computational studies that support the original hypothesis. We are also investigating how these structural changes lead to changes in substrate selectivity.
Some results from our work!
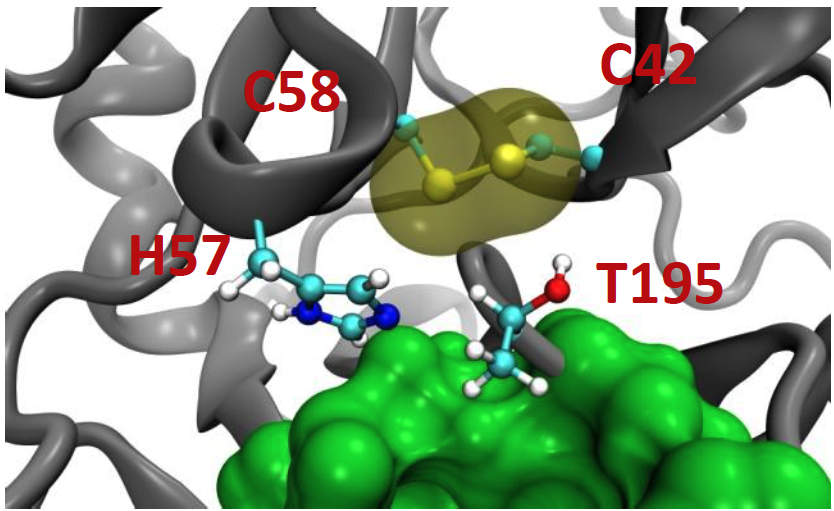
From modeling studies carried out by the Guliaev let, we can see that the hydroxyl group of Thr In the S195T substitution does not hydrogen bond with His57. We hypothesized that the disulfide bridge formed by residues 42 and 58 preclude T195 from achieving the conformation that is required for hydrogen bonding between the side chains.
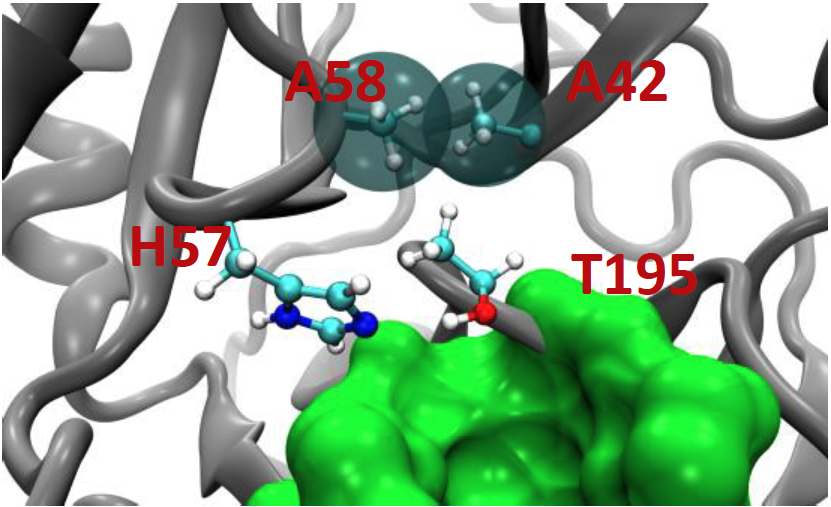
C42 and C58 replaced with alanine, the hydroxy group of T195 is now capable of hydrogen bonding with His 57! This interaction is not permanent, however. The structural stability that is lost by the removal of the C42-C58 disulfide bridge allows the side chain of T195 to rotate and occupy two other inactive conformations.
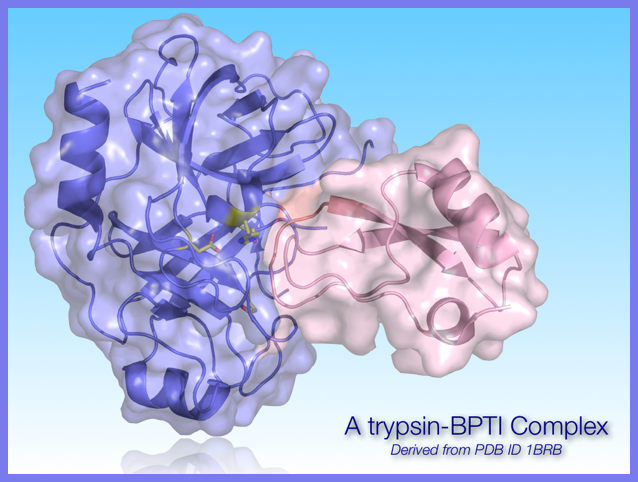
Proteases are involved in nearly every biological process in some way. Consequently, they have long been targets of small molecule drugs. However, more recently, proteases (particularly trypsin-fold proteases) are now being developed to be used as drugs themselves. One of the inherent difficulties is the presence of naturally occurring inhibitors that exist to prevent unnecessary or errant proteolysis. Once a proteolytic drug is delivered, its lifetime is relatively short because of such protective measures. Our lab is using protein engineering approaches to identify sites that are important for protease-inhibitor association with the intent of weakening these interactions without eliminating or reducing catalytic activity significantly.
Some results from our work!
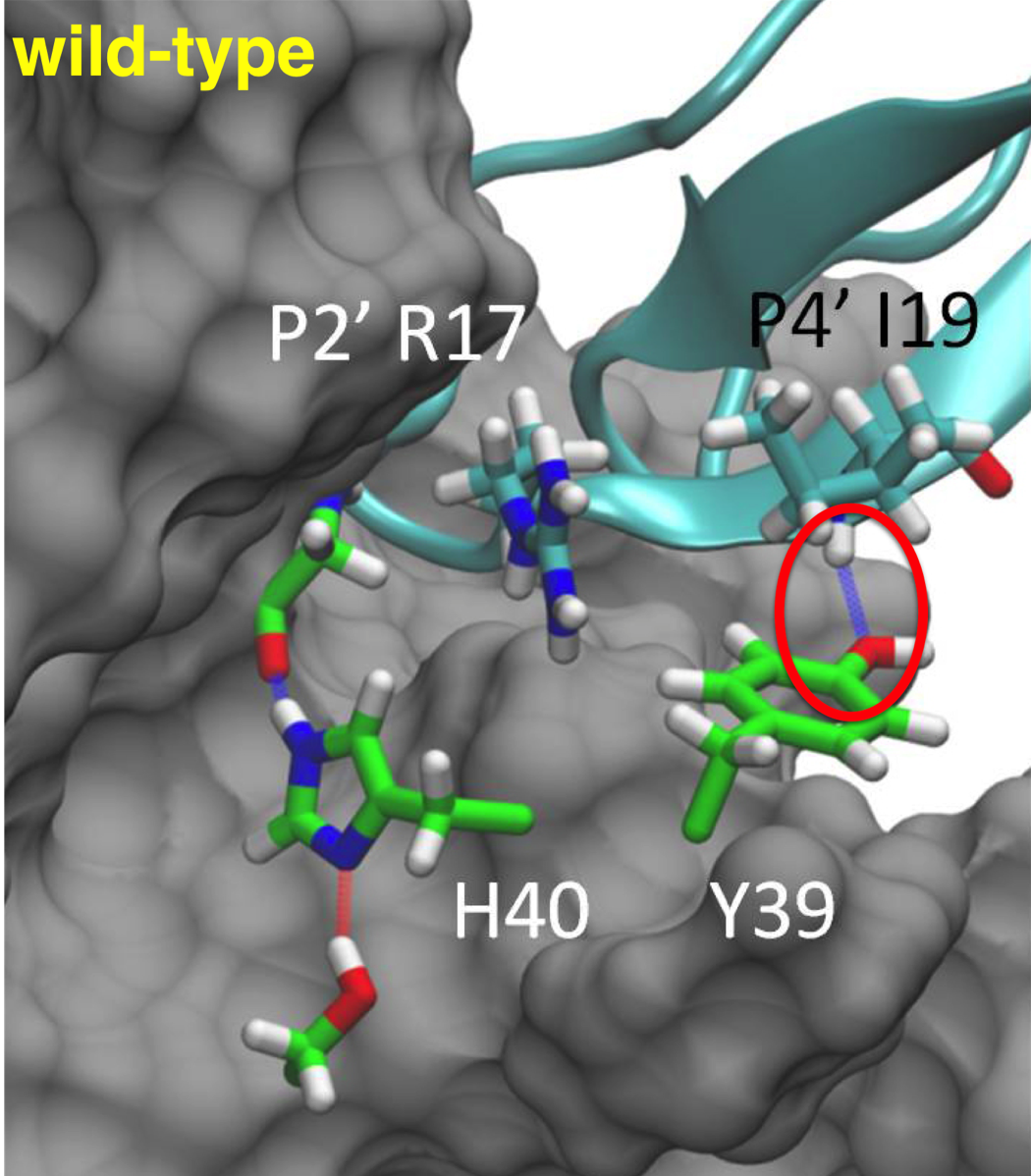
In wild-type trypsin, the P4' backbone amide of the inhibitor hydrogen bonds with the hydroxy group of Try39 in trypsin.
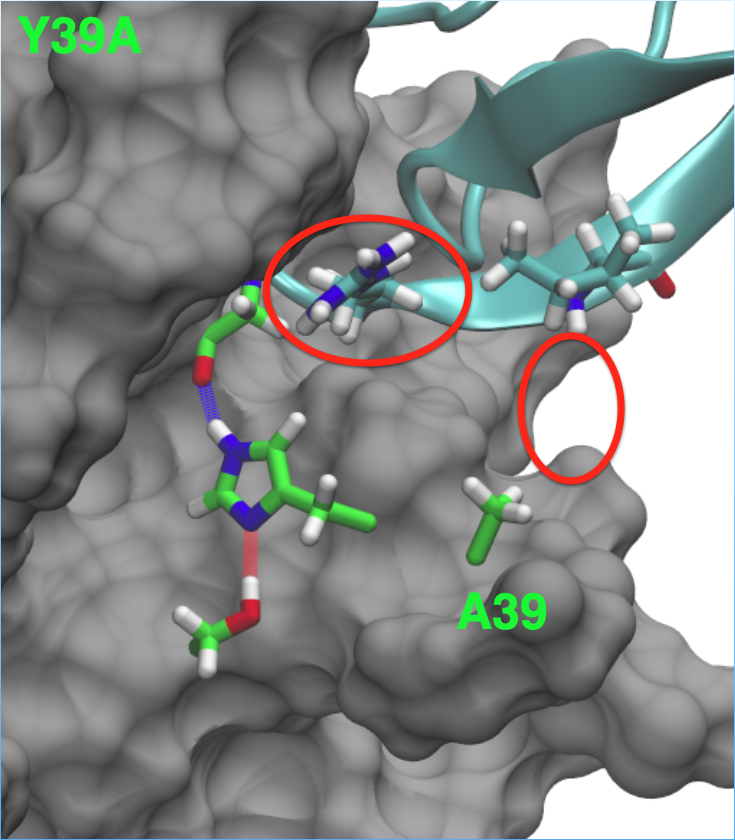
The Y39A substitution removes this hydrogen bonding capability. The result is a decrease in affinity for the inhibitor. The substitution does not reduce the activity of the enzyme. This was a very promising result!
Information & Resources
Contact
Teaster Baird, Jr., Department Chair / Professor
Phone: (415) 338-1288
Email: tbaird@sfsu.edu
Lab Location
Thornton Hall, Room 815
Office Location
Thornton Hall, Room 806
Office Hours
Tuesdays, 3 p.m. - 5 p.m.
Fridays, 10 a.m. - 12 p.m.
Virtual Office Hours - Spring 2021
Due to restrictions that have been implemented to combat COVID-19, all office hours are virtual but will still be held at the same times via Zoom. For more information please send email to tbaird@sfsu.edu.
Publications
Batt, A.R., St. Germain, C.P., Gokey, T., Guliaev, A.B., Baird, T. Jr., Engineering trypsin for inhibitor resistance. Protein Science (2015) DOI: 10.002/pro. 2732
Tansey, J.T., Baird, T. Jr., Cox, M. M., Fox, K.M., Knight, J., Sears, D., Bell, E. Foundational concepts and underlying theories for majors in Biochemistry and Molecular Biology Biochem. Mol. Biol. Educ. (2013) 41, 287-296
Baird, T. T., Jr., and Craik, C. S. Trypsin Handbook of Proteolytic Enzymes, 3rd ed. (2013) 2594-2600
Baird, T. T., Jr., Trypsin, Brenner's Encyclopedia of Genetics, 2nd edition (2013) pp. 216-219
Gokey, T., Baird, T.T., Jr., and Guliaev, A.B. Conformational dynamics of threonine 195 and the S1 subsite in functional trypsin variants J. Mol. Model (2012) 18, 4941-4954
Baird, T.T., Jr., Wright, W. D., and Craik, C. S. Conversion of Trypsin to a Functional Threonine Protease, Prot. Sci., (2006) 15, 1229-1238.
Halfon, S., Baird, T.T., Jr., and Craik, C. S. Trypsin Handbook of Proteolytic Enzymes, 2nd ed. (2004) 1483-1488
Baird, T.T., Jr. and Craik, C. S. Trypsin. Encyclopedia of Genetics, Academic Press, London (2001) 2071-2075.
Baird, T. T., Jr., Wang, B., Lodder, M., Hecht, S. M., and Craik, C. S. Generation of Active Trypsin with Chemical Cleavage, Tetrahedron Letters (2000) 56(48), 9477-9485
Wang, B., Lodder, M., Baird, T. T., Jr., Brown, K. C., Craik, C. S., and Hecht, S. M. Chemically Mediated Site-Specific Cleavage of Proteins, J. Am. Chem. Soc. (2000) 122, 7402-7403
Doyon, J. B., Hansen, E. A. M., Kim, C.-Y., Chang, J. S., Christianson, D. W., Madder, R. D., Voet, J. G., Baird,T. T., Jr., Fierke, C. A. and Jain, A. Linear Free Energy Relationships Implicate Three Modes of Binding for Fluoroaromatic Inhibitors to a Mutant of Carbonic Anhydrase II, Organic Lett. (2000) 2, 1189-1192.
Alber, B. E., Colangelo, C. M., Dong, J., Stålhabdske, C. M. V., Baird, T. T., Tu, C., Fierke, C. A., Silverman, D. N., Scott, R. A. and Ferry, J. G. Kinetic and Spectroscopic Characterization of the Gamma Carbonic Anhydrase from the Methanoarchaeon Methanosarcina thermophila, Biochemistry (1999) 38, pp. 13119-28
Baird, T. T., Jr., Waheed, A., Sly, W. S., and Fierke, C. A. Catalysis and Inhibition of Human Carbonic Anhydrase IV, Biochemistry, (1997) 36 pp. 2669-2678
Ippolito, J.A., Baird, T.T., Jr., McGee, S. A., Christianson, D. W., and Fierke, C. A. Structure-assisted redesign of a protein-zinc-binding site with femtomolar affinity, Proc. Natl. Acad. Sci. USA, (1995), 92 pp. 5017-5021.
Wild, C., Dubay, J. W., Greenwell, T., Baird, T., Jr., Oas, T. G., McDanal, C., Hunter, E., and Matthews, T. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc. Natl. Acad. Sci. USA, (1994), 91 pp. 12676-12680
Miller, T. M., Kwock, E. W., Baird, T. Jr., and Hale, A. Properties and Photodefinition of Poly(aromatic diacetylenes). MATERIALS, (1994) 6 pp. 1569-1574
Kwock, E. W., Baird, T., Jr., and Miller, T. M. Synthesis and Characterization of Soluble, High Molecular Weight Poly(aromatic diacetylenes), Macromolecules, (1993), 26 pp. 2935-2940
Courses Taught
Undergraduate Biochemistry Courses
CHEM 340: Biochemistry I
- Protein structure and function; carbohydrate metabolism; enzyme kinetics and thermodynamics; electron transport chain.
- Spring 2021 Syllabus (COMING SOON!)
CHEM 341: Biochemistry II
- Biosynthesis and degradation of lipids, amino acids, and nucleotides; protein synthesis, folding and degradation; DNA and RNA structure; DNA replication and transcription.
- Fall 2020 Syllabus
CHEM 343: Biochemistry I Lab
- Chemical and enzymatic experiments involving proteins, carbohydrates, and nucleic acids with emphasis on enzyme kinetics. Utilization of standard and modern biochemical techniques.
- Fall 2016 Syllabus
CHEM 349: General Biochemistry
- Survey of major areas in biochemistry including enzymology, bioenergetics, and carbohydrate, lipid and nucleic acid metabolism.
Other Undergraduate Courses
CHEM 115: General Chemistry I
- Essential concepts of atomic properties, atomic interactions, reaction chemistry, stoichiometry, thermodynamics, chemical kinetics, and equilibria.
CHEM 470: Biochemistry Research
- Intended for motivated students interested in independent research. Requires 6-9 hours of research each week, lecture attendance, formal poster presentation, and written report
CHEM 699: Independent Research in Biochemistry
- Laboratory or library research work focused on chemistry and biochemistry problems directed by a department faculty. For advanced, superior students majoring or minoring in chemistry or biochemistry.
Graduate Courses
CHEM 800: Proteins and Enzymes: Structure, Function and Engineering
- Understanding structure-function relationships in proteins and enzymes.
CHEM 880: Graduate Seminar
- Exploration of current areas and methods of chemical research. Oral presentations and scientific writing. Should be taken during the first year of graduate work.
Podcasts - Links Coming Soon
01: Water-Introduction
January 5, 2020
You may think you don't need an introduction to water, but maybe you don't know it as well as you think you do. (Recorded spring 2019)
Time - 3:52
02: Water-Physical Properties
January 5, 2021
The freakiness of water as a chemical arises from its physical properties. Let's look closely at it. (Recorded spring 2019)
Time - 17:11
03: Hydrogen Bonding
January 5, 2021
You can call me bond…hydrogen bond. (Previously recorded)
Time - 7:53
04: Water as a Solvent
January 5, 2021
We are bags of water with stuff dissolved in it. How does that work? (Previously recorded)
Time - 14:14
05: Water and non-polar Solutes
January 5, 2021
Water is a very polar substance, so how does it deal with hydrophobic things? (Previously recorded)
Time - 9:30
06: Weak Acids
January 5, 2021
Not the strong stuff, just a little review on weak acids. (Previously recorded)
Time - 16:32
07: Amino Acid Titrations
January 5, 2021
Amino acids are also weak acids and they behave as such. Let's consider how a titration of a weak acid may look. (Previously recorded)
Time - 14:25
08: pH and pKa Relationships
January 5, 2021
Like any other weak acid, the pKa values of an amino acid's titratable groups provides valuable information. (Previously recorded)
Time - 9:14
09: Calculation of Net Charge
January 5, 2021
Amino acids are never uncharged in solution. What's cool is that we can calculate to what degree it will be charged if we know the pKa values for the amino acid and the pH of the solution. (Previously recorded)
Time - 14:55
10: Calculation of pI
January 5, 2021
Even though amino acids are never uncharged in aqueous solution, there can be a "net neutrality." (Previously recorded)
Time - 16:39
11: Peptides
January 5, 2021
Ok, we've dealt a lot with amino acids. However, they can be joined to make peptides. Let's talk about them. (Previously recorded)
Time - 13:02
12: Protein Structure-Introduction
January 5, 2021
Now we can move on to the "big" stuff. Let's prime ourselves to talk about protein structure.(Previously recorded)
Time - 5:35
13: Protein Secondary Structure
January 5, 2021
A protein without a defined structure, doesn't have a defined function. Let's talk about how structure is defined in proteins. (Previously recorded)
Time - 2:18
14: Protein Tertiary Structure
January 5, 2021
Coming soon.
Louis Stokes Alliance for Minority Participation (LSAMP)
The California State University Louis Stokes Alliance for Minority Participation (CSU-LSAMP) is a CSU wide academic support program that is supported by funds from the National Science Foundation and the CSU Chancellor's office. As a Professional Preparation Emphasis program, the goals of LSAMP at SF State are to 1) increase the number of students who graduate with science, technology, engineering and math (STEM) degrees, targeting those who face or have faced social, educational or economic barriers to careers in STEM and 2) to provide resources to make such students more competitive applicants to STEM PhD programs.
In the LSAMP program SF State, we employ several strategies to meet this goal including:
- A Textbook Reimbursement Program*: LSAMP participants can receive partial reimbursement for eligible textbooks each academic year.
- Research Support*: LSAMP participants who are conducting research with an SF State STEM faculty member can receive a stipend to help in research activities and attending scientific conferences.
- Professional Training*: LSAMP participants can also participate in a series of workshops focused on developing skills that will better prepare you for a research career. The workshops focus on topics such as communicating your science to various audiences, developing presentation skills, learning how to read the scientific literature--all skills anyone who aspires to be a successful scientist will have to master.
- Other opportunities*: There are also opportunities available through the CSU state-wide network.
*Restrictions and limitations apply. Please contact Dr. Baird, the SF State Campus Coordinator, for details.
How to Apply
Eligibility
In order to be considered for the LSAMP program, a student must:
- be a U.S. citizen or permanent resident
- be an undergraduate enrolled at SF State (the program does not fund graduate students)
- be a declared STEM major with an overall GPA of 2.5 or better
- be an individual who has faced or faces social, educational, or economic barriers to careers in STEM
- complete a 2020 - 2021 CSU-LSAMP application (The 2020 - 2021 application is a fillable PDF. Please download it and type in your responses. The 2010 - 2020 application has been discontinued)
Forms
- Application (PDF)
A current LSAMP Application must be submitted and approved before one can take advantage of LSAMP benefits. - Stipend Application (PDF)
After LSAMP Application approval, LSAMP participants are eligible to receive financial support for research projects that are supervised by SF State faculty. Complete the stipend application for this benefit. - Reimbursement Request (PDF)
LSAMP participants are eligible to be reimbursed for textbooks, partial travel, conference fees and GRE fees. Use the Reimbursement Request to request this support. Reimbursement requests must be accompanied by original receipts.
Submit
After completing the application, stipend and/or reimbursement request form(s), please email the document(s) to Dr. Baird tbaird@sfsu.edu or print it and take it to Dr. Baird's office inside TH 806.
CSU-LSAMP is funded through the National Science Foundation (NSF) under grant #HRD-1826490 and the Chancellor's Office of the California State University. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation or the Chancellor's Office of the CSU.
Other Links
Protein Structure
- The Protein Data Bank
- Structural Classification of Proteins
- ExPASy Bioinformatics Resource Portal
- PyMOL
- Rasmol
- Swiss PDB Viewer
- Protein Explorer Options
Professional Societies and Organizations
- American Society for Biochemistry and Molecular Biochemistry (ASBMB)
- ASBMB Student Chapters
- The American Association for the Advancement of Science (AAAS)
- The Protein Society
Journals
- Journal of Biological Chemistry (JBC)
- Science
- Nature
- Biochemistry
- Journal of the American Chemical Society
- Proceedings of the National Academies of Science (PNAS)
- Protein Science
- Journal of Molecular Biology (JMB)
- Biochemistry and Molecular Biology Education (BAMBED)
Other Useful Sites

About the PI
I was born and raised in Ashland, Mississippi, a small town of 500 people. There wasn't much to do there so I decided that I would go to college when I finished high school. After high school, I attended Tougaloo College in Tougaloo, MS where I earned a BS in chemistry. It was there that I got my first taste of scientific research and lab work through the MARC program.
The MARC program allowed me to have research experiences in biochemistry at Purdue University and bioanalytical chemistry at Eli Lilly and Company. While at Tougaloo, I worked in the laboratory of Dr. Mona Trempe in the Department of Biochemistry at the University of Mississippi Medical Center in Jackson, Mississippi where I did biophysical studies on an aminoacyl tRNA synthetase complex.
My research experiences caused me to develop a fondness for biochemistry, so from Tougaloo, I went to graduate school in biochemistry at Duke University in Durham, NC. There, I studied mechanistic enzymology under the guidance of Dr. Carol A. Fierke (now Provost at Brandeis University). I worked with two isozymes of carbonic anhydrase, CA II and IV where I characterized CA IV and did some protein engineering with CA II.
I really liked the protein engineering aspect of my graduate work, so after I graduated from Duke, I did a post-doc with Dr. Charles Craik at the University of California, San Francisco where I continued work that he had begun with the serine protease, trypsin.
After a few years there, I was fortunate enough to secure a position at San Francisco State University where I am today and I continue to work with trypsin. I am also the current Chair of the Department of Chemistry & Biochemistry at SF State.
Download Adobe Reader to view the PDF files on this page.
